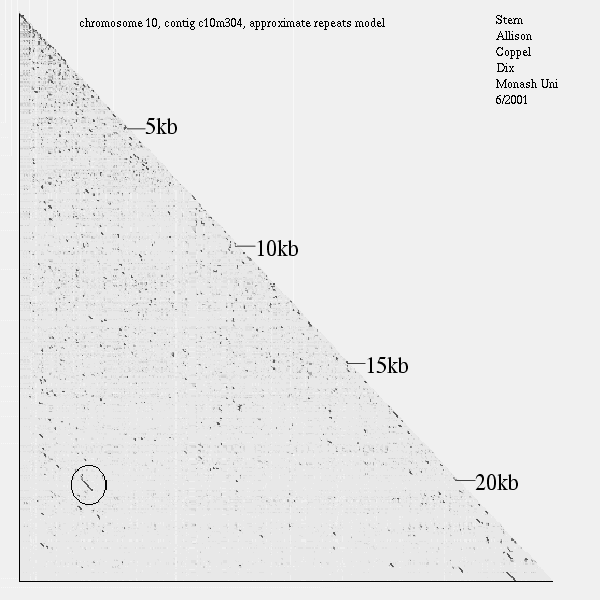
| alphabetic order |
:Bioinformatics
|
|---|
| track record |
interest |
useful technology, e.g. |
|---|
|
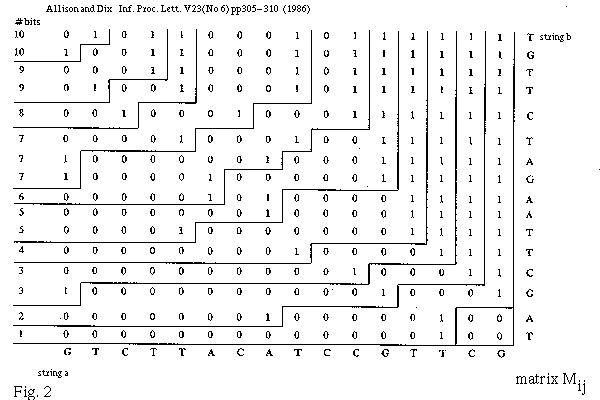
IPL.V23 1986...
| * |
MML, algorithms, data mining
|
|---|
|
* | algorithms, parallel sys', data mining |
|---|
David Dowe |
HICSS-26 1993...
| * | MML, Stats |
|---|
|
| * | constraints, optimization |
|---|
|
J.Theor.Biol.V23 1969...
| * |
MML, inductive inference, clustering |
|---|
This
document can be found at
users.monash.edu.au/~lloyd/Seminars/2001-VBC01/index.shtml
and contains hyper-links to other resources
- © L. Allison, 7 September 2001.

|
>>programme>>

|
In: SOBS Research Linkage Seminars,
Friday September 7th 2001
- 3.00 - 3.10
- Dr. Brian Cooke and A/Prof. John Davies.
Dept. Microbiology, Monash University.
SOBS Research Linkage Seminars - introduction and overview.
- Session 1 Genomes (Chair: A/Prof. John Davies)
- 3.10 - 3.25 Prof. Ross Coppel.
Dept. Microbiology, Monash University.
Genomes. Why are they important?
- 3.25 - 3.40
- Dr. Sue Forrest. Australian Genome Research Facility.
Facilitating Genome Research - inside the AGRF.
- 3.40 - 3.55
- Prof. Ross Coppel. Dept. Microbiology, Monash University.
The Victorian Bioinformatics Consortium (VBC).
- 3.55 - 4.10
- Dr. Lloyd Allison.
School of Computer Science and Software Engineering, Monash University.
Bioinformatics @ CSSE (left).
- Break
- Session 2 Immediate post-genome (Chair: Dr. Brian Cooke)
- 4.30 - 4.45
- A/Prof. John Davies. Dept. Microbiology, Monash University.
A DNA core facility. Oligos, sequencing and robotics.
- 4.45 - 5.00
- Dr. John Boyce. Dept. Microbiology, Monash University.
Real-time PCR; accurate quantitation at the speed of light.
- 5.00 - 5.15
- A/Prof. John Davies. Dept. Microbiology, Monash University.
Microarrays.
- 5.15 - 5.30
- Dr. James Whisstock.
Dept. Biochemistry and Molecular Biology, Monash University.
Utilising genomic data.
- 5.30 - 5.35
- Prof. Warwick Anderson.
Dept. Physiology, Monash University.
Genomes and beyond.

|